Biol 240H Course Blog
Posts will appear here for your own interest. They will not be covered on exams.
Tuesday, February 24, 2026
Curiosity creates cures
Thursday, February 19, 2026
Cytoskeletal dynamics movies
- See beads whiz around: Actin comets and motility induced by an activator of an actin nucleator stuck onto beads
- A GFP-tagged protein that binds only to the plus ends of growing microtubules
- Beautiful movies of cells using a microscopy trick that we didn't talk about in the course (lighting up cells with a super-thin sheet of light coming in from the side) and tumor cells expressing a GFP-tagged polypeptide that binds actin filaments, viewed like this
Wednesday, February 18, 2026
Cystic fibrosis and drug development
Trikafta is a 3-part drug that helps deltaF508 cystic fibrosis patients. Trikafta was approved by the FDA in 2019 based on studies testing efficacy and safety [1],[2]. The figure below illustrates mechanisms of action for a previous 2-part drug and the new 3-part drug.
Bad news though: the drug costs more than $300,000 per year (so research on more cost-effective solutions is critically important). Developing drugs like these costs a lot, and drug companies need to recoup costs and make a profit to justify investing in possible new drugs, so the FDA gives companies a 5-7 year monopoly when a drug is developed, before generic versions of drugs can be sold. The complex issues involved are covered well, I think, here.
You can find info about cystic fibrosis from the NIH, and current clinical trials here.
Here's an old but really interesting article about Cystic Fibrosis, and a source for up-to-date information on the disease and its effects on patients.
Tuesday, February 17, 2026
Kizzmekia Corbett, a Tar Heel and a science hero

Among the scientists we have to thank for the COVID vaccines that were transformational in the pandemic is one person who played a central role in the development of RNA-based vaccines, Dr. Kizzmekia Corbett.
Dr. Corbett grew up near Chapel Hill, worked in a UNC lab for a summer in high school, and then moved back and forth between North Carolina and the DC/Baltimore area as a researcher: she was a student in the famous Meyerhoff Scholars Program at UMBC, then did her PhD at Carolina, and then moved to the National Institutes of Health.
At the NIH, Corbett became the scientific lead of the Vaccine Research Center's COVID-19 team that developed mRNA-based vaccines by making use of the Cryo-EM structure and her and colleague's previous work on MERS virus to design an mRNA vaccine rapidly. Corbett and colleagues discovered that the mRNA vaccines worked in mice and in primates, leading to the Moderna and Pfizer mRNA vaccines that transformed the pandemic.
 |
| December 2020: The efficacy of the Moderna (see p. 30) and Pfizer (see p. 58) mRNA vaccines was shockingly good! Whew! |
Now she's a professor at Harvard, running a lab that studies vaccines and the immune system, to inform the development of new vaccines including (fingers crossed) a vaccine that might work against all SARS-COV-2 virus variants. She's been active on social media, which has given people an almost-live, inside view of what must be a fascinating path.
Here's a general-interest interview with then-NIH chief Francis Collins...
... and I recommend especially another interview that spans from her initial interest in science to some more in-depth virology:
Thursday, February 12, 2026
Try your hand at some simple bioinformatics
To start with, you might try a program that can predict the arrangement of transmembrane proteins based on sequence. The program, described in more detail than most people will want to see here, uses rules deduced by biologists, and was then fine-tuned with an artificial intelligence algorithm, training the program using transmembrane proteins with known orientations. There is a similar program that can predict where in a cell a protein will end up.
Then you can try other predictions that can be made based solely on sequence by searching online for other predictor tools.
(2) See how little a well-conserved protein has changed through evolution. Let's look at the current versions of beta-tubulin from yeast and human, and see how similar they are. Try this: copy the text of the yeast beta-tubulin sequence from here (from MREI... to the end of the protein's sequence), then paste it into the Enter Query Sequence box here, under Choose Search Set, for Database choose non-redundant protein sequences (nr) and next to Organism, type "human" and then select human from the dropdown menu that appears ('human' or 'humans'). Then click BLAST at the bottom. The page will automatically update for seconds or minutes, depending on how busy servers are. When it's done, you'll see the results in a tab marked Alignments. You'll see the sequence you queried (the yeast beta-tubulin) and the subject sequence – the closest protein sequence that it could find among all known human proteins. In between is a list of identical amino acids, and + signs for similar amino acids (similar based on charge, etc). Tubulin, actin, and histones are remarkably well conserved proteins – they've had very few changes across hundreds of millions of years of evolution. If you try the exercise with other kinds of proteins, you'll see that only parts of them are well conserved across diverse organisms, or that some don't exist in certain organisms.
More about Bioinformatics at Wikipedia.
(image: a 7-transmembrane protein from Wikimedia Commons)
Wednesday, February 11, 2026
Nobel Prizes
 |
| UNC's Aziz Sancar on a Turkish postage stamp |
The Nobel Prize web site has great, clear explanations of the important discoveries that have been recognized with the award, along with the key experiments that led to these discoveries. The newest ones have both introductory and advanced explanations as pdf files.
The Nobel Prizes are announced each October. Most of the ones below were awarded for discoveries that we're discussing in class.
- 1974: George Palade and the secretory process
- 1986: Cell-cell signaling by growth factors
1992: Reversible protein phosphorylation as a biological switch
Good spot for a selfie!
Smithies' old UNC parking
spot on Medical Drive- 1997: Certain proteins (prions) can act as infectious agents
- 1999: Discovery of signal sequences for protein sorting
- 2001: The cell cycle
- 2007: Oliver Smithies at UNC! (Oliver passed away in 2017).
- 2008: GFP
- 2012: G protein-coupled receptors in cell communication
- 2014: Super-resolution microscopy
- 2015: UNC's Aziz Sancar for DNA repair mechanisms! Go Heels!
- 2017: CryoEM
- 2020: CRISPR - Jennifer Doudna (pictured below) will be speaking at UNC on March 26. If you want to see her and hear the talk, go early for a good seat!
 |
| Charpentier and Doudna, from a great video about their work on CRISPR and their 2020 Nobel Prize. |
Tuesday, February 10, 2026
Turning a molecular wheel with a flow of protons
 |
| (image source) |
A scientist's story of first hearing Peter Mitchell's proposal that a concentration gradient of protons was the energy source for ATP synthase.
I recommend reading this starting in the middle, with "In 1955..." (highlighted, second sentence of the 3rd paragraph) and then looping back to the beginning of the article afterwards.
"I remember thinking... that I would bet anything that ATP synthesis didn’t work that way."Mitchell was an unusual scientist:
"For much of his career he worked in his own lab in a... house... his research funded in part by a herd of dairy cows." (source)
Sunday, February 8, 2026
Thursday, January 29, 2026
Sickle cell anemia
Learn more about sickle cell anemia, the most common inherited blood disorder in the US, at an NIH web site or the Sickle Cell Disease Association of America web site.
Here's a NY Times story about recently approved gene therapy (for those interested in science writing, the author Gina Kolata is a great writer who tells engaging stories while reliably getting the science right, and she has several books out). Gene therapy is an important step forward, but sadly, as this story covers, the current cost of gene therapy makes it unavailable to many patients, and as these stories cover, gene therapy patients have faced some challenges to date.
Here's an earlier 22-minute radio story about the first US patient whose bone marrow cells' genes were edited using CRISPR.
Get an idea of what else is being tried right now toward cures and toward treatments for symptoms by seeing current clinical trials.
Wednesday, January 28, 2026
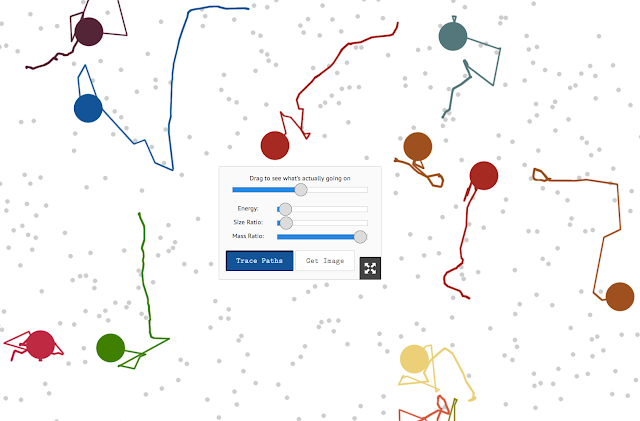
Brownian motion simulation
Tuesday, January 27, 2026
Dorothy Crowfoot Hodgkin
(Image: British postage stamp honoring Hodgkin)
At the beginning of the course, I explained that the science we are discussing intersects with other aspects of life in many ways, and that I'd discuss those ways on occasion. For anyone interested in how women have been portrayed in science journalism, here's a headline from when Hodgkin won the Nobel Prize, and another, plus a newer checklist developed to help science journalists think clearly about whether their writing rehashes cliches at the expense of explaining the science and the scientist's achievements. Here you can read a little detail about the exclusion of women in research discussions in the time and place where Hodgkin was making discoveries.
Monday, January 26, 2026
How many diseases are linked to specific genes? How many disease genes are there?
 |
| (image credit) |
- How many diseases have genes associated with their malfunctions? Here is a periodically-updated list. As of 15 years ago (Jan 27, 2011), 2949 diseases were known with "Phenotype description, molecular basis known: Total". Today (Jan 26, 2026) it's 7091 diseases – an average increase of nearly a new one discovered per day!
- How many disease genes are there? See "Total number of genes with phenotype-causing mutation" here. As of today (Jan 26, 2026) it's 5045 genes.
Saturday, January 24, 2026
The earliest snowflake images
In 1885, Vermont farmer Wilson Bentley attached a camera to a microscope and became the first person to successfully photograph an individual snowflake crystal.
"...it was my mother who made it possible for me, at fifteen, to begin the work to which I have devoted my life. She had a small microscope which she had used in her school teaching. When the other boys of my age were playing with popguns and slingshots, I was absorbed in studying things under this microscope: drops of water, tiny fragments of stone, a feather dropped from a bird's wing, a delicately veined petal from some flower. But always, from the very beginning, it was the snowflakes that fascinated me most." -from a 1925 interview with Bentley
Thursday, January 22, 2026
Henrietta Lacks
About the book about Lacks that I mentioned in class: "...the science end of this story is enough to blow one's mind... But what's truly remarkable... is that we also get the rest of the story, the part that could have easily remained hidden had she not spent ten years unearthing it: Who was Henrietta Lacks? How did she live? How she did die? Did her family know that she'd become, in some sense, immortal, and how did that affect them?" -Jad Abumrad, Radiolab
There's a movie too.
Tuesday, January 20, 2026
Saturday, January 17, 2026
Amazing images from microscopes
Friday, January 16, 2026
Breaking the resolution limit of light microscopy
 |
| image credit: HHMI |
"...that’s where we learned about photoactivatable fluorescent proteins.... it became obvious to Harald and me that this was the missing link for the idea that I had pitched after I left Bell: we could isolate a few molecules at a time by activating limited subsets of photoactivatable proteins. It seemed so easy."
"...We were both unemployed, but Harald had some of his equipment from Bell [Labs]..."
-from Betzig's Nobel speech
Xiaowei Zhuang's lab developed a similar method, called STORM, around the same time as PALM was developed. See "STORM image gallery" at her lab web site for some super-resolution images.
Thursday, January 15, 2026
Struggling with your grades? Fear not!
Watch the 45 seconds between 1:45 and 2:30 to see how Chemistry Nobel Laureate Marty Chalfie (of Green Fluorescent Protein fame) did in Chemistry classes in college.
Wednesday, January 14, 2026
Louis Pasteur and his home
Louis Pasteur's old house is now a museum, currently under renovation through 2028. You can see it well documented here. Pasteur is buried in the basement, in an elaborate crypt (pictured here).
Tuesday, January 13, 2026
Microns, nanometers, etc.
Powers of Ten, a classic Charles & Ray-Bernice Eames film on grasping the sizes of things from human scale out to the then-known universe, and back in to subatomic particles.
Friday, January 9, 2026
More images from Robert Hooke's Micrographia
Here's the whole book.
Hooke's book begins with an apology to the King of England, for, well, discovering things.
Hooke's methods for immobilizing some of the insects were creative:
...I gave it a Gill of Brandy, or Spirit of Wine, which after a while e'en knock'd him down dead drunk, so that he became moveless...
 |
| Hooke's engraving of a head of a hoverfly, from Micrographia |
Thursday, January 8, 2026
My favorite of van Leeuwenhoek's letters, with a surprise ending
In this letter, he describes finding microscopic life in frog poop. I especially like the surprise ending in the last 3 paragraphs – starting, "On the fifth day the Frog had dung'd again...." on the bottom of page 517. Scientific papers never end like this.
 |
| Google on van Leeuwenhoek's 384th birthday |
Wednesday, December 17, 2025
A popular book about cell biology
A great, vivid exploration of some fascinating topics in cell biology. See the "Read sample" button on the left side of this page to read parts of the book now. Also available as an audiobook.